
PURPOSE: Apoptosis is an important process used by cells to invoke their own demise and vitally important for growth and regulating cancer. A key protein responsible for ensuring that this process occurs at the right time is BclXL Colleague Vikas Bhat has dedicated his doctoral thesis to the study of the behavior of this protein, in other words, how its molecular structure changes in different conditions. The purpose of the work to be shown here is to explore the effects of acids and bases on this protein.
HYPOTHESIS: Specific amino acids are susceptible to changes in pH of the buffer and can switch from being charged to becoming neutral. Such changes in turn might break positive-negative type bonding, yet promote different types of bonding leading to rearrangement of structure. As BclXL protein has a significant amount of acidic amino acids that exist in key positions able to affect the overall structure of the protein, it is the hypothesis of this experiment that pH changes will have dramatic effects.
METHODS: The work shown requires isolated protein that is then analyzed using two amazing techniques. To isolate protein, the DNA the encodes the protein is transformed into E. Coli bacteria. The bacteria cells create the protein and then are split open to recover the protein lysate. This is then run on purification columns to extract the BclXL protein. From here the protein in changed into solutions that have pH set to either 4, 6, 8 or 10 and then analyzed with the first technique analytical light scattering. This technique hits the protein in solution and catches light the protein scatters at 3 different angles. Using the intensities of light it collects from these angles the molecular weight of the protein can be determined. Finally, the protein is then dried onto special slides to be observed using scanning electron microscopy. This technique magnifies the protein 50,000 times which allows for observe large bundles of protein if they form.
RESULTS: A single BclXL monomer has the potential to bind together to create a dimer or combine into larger structures called multimers, polymers or megamers. Information on how pH can affect formation of these structures can be determined using analytical light scattering (Figure 1a). The data uncovered here shows that extremely large structures form at the most acidic pH and BclXL forms the smallest structures at pH 6. Figure 1b demonstrates the values coming from the three detectors of scattered light and a linear fit of the line allows for calculation of the molecular weights. BclXL monomer is approximately 31 kilodaltons (kDa) while the analytical light scattering detects that at pH 4.0 this protein comes together to form ~34,000 kDa sized structures. Additionally, the multimer is roughly 300-500 kDa and the polymers ~3000-3500 kDa.
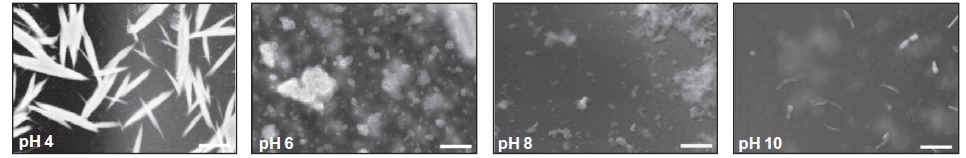
If indeed structures of this size exist then they may be observable using scanning electron microscopy, as shown in Figure 2. Indeed as predicted by the light scattering the structures in the BclXL mixture at pH 4.0 have formed into extremely large rod like structures and these structures break apart at higher pH.
FIGURE 1: Analytical light scattering analysis of BclXL free protein at different pH.
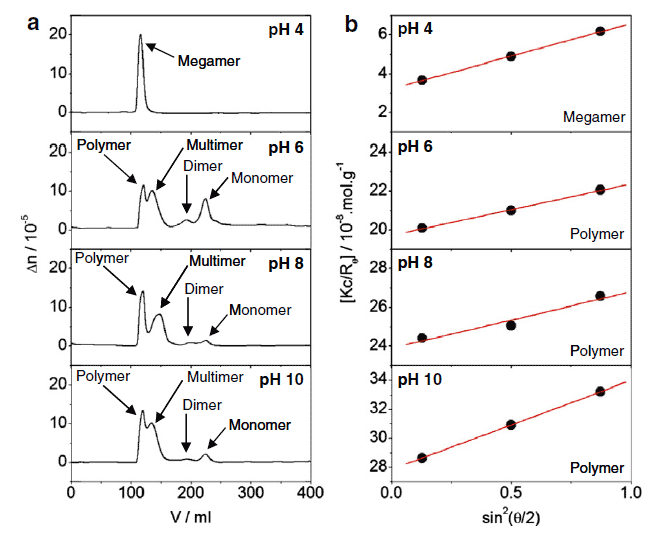
FIGURE 2: Highly magnified (50000x) image of BclXL protein using a scanning electron microscope.
DISCUSSION: The work described here is just two of many experiments performed in the study that explore how pH affects the behavior of proteins. These data alone give insight into how at pH 6.0 the protein exist mostly in its single monomeric form and ready to bind to other proteins to perform its function, while at pH higher and lower than 6.0 the protein binds to itself to keep from performing its function. Such behavior is well suited to observations of large pH changes in cells during apoptosis and how this may in fact be used as a tool to change cell behavior. The rest of this work can be seen either here (pubmed) or here (FarooqLab > Publications > Research Papers > #51). ScientificAmeriKen would like to thank Dr. Bhat et al. for the great work here as well as for the fancy pictures.